Unveiling the fragility of language models to drug names
RABBITS: A new medical robustness investigation and LLM benchmark
July 19, 2024
Written byBittermanLab
Motivation and Problem
Language models (LLMs) like GPT are touted as game-changers in the medical field, providing support in data processing and decision-making. However, there's a significant challenge: these models struggle with the variability in drug names. Patients often use brand names (like Tylenol) instead of generic equivalents (like acetaminophen), and this can confuse LLMs, leading to decreased accuracy and potential misinformation. This is a critical issue in healthcare, where factuality is paramount.
What We Did
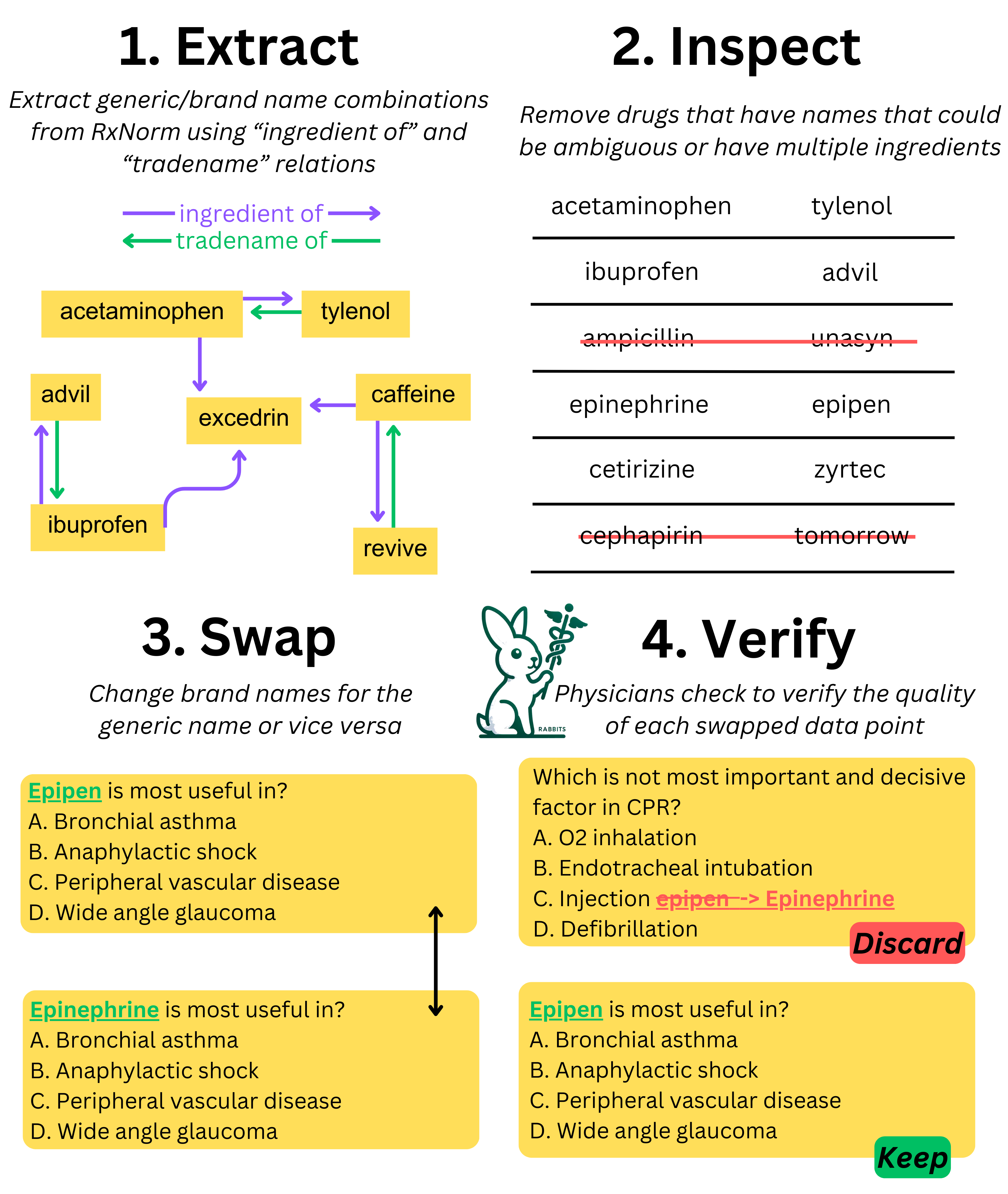
To tackle this problem, we developed a specialized dataset called RABBITS (Robust Assessment of Biomedical Benchmarks Involving drug Term Substitutions for Language Models). Here's what we did:
1. Dataset Creation: We used the RxNorm database to generate a comprehensive list of brand and generic drug pairs. This involved identifying 2,271 generic drugs and mapping them to 6,961 brands.
2. Data Transformation: Using regular expressions, we created two versions of medical QA datasets (MedQA and MedMCQA): one with brand names swapped to generics and one with generics swapped to brand names.
3. Expert Review: The transformed datasets were rigorously reviewed by physician experts to ensure accuracy and context consistency.
4. Evaluation: We evaluated various open-source and API-based LLMs on these transformed datasets using the EleutherAI lm-evaluation harness in a zero-shot setting. The goal was to measure performance differences when drug names were swapped.
What We Found
We found that language models answer medical questions differently when brand vs. generic drug names are used. There was a consistent performance drop ranging from 1-10% on our benchmarks. Why does this happen?
1. Data Contamination: One major factor is that LLMs are often trained on datasets that include test data (Over 90% of MedQA questions appeared to some extent in Dolma Dataset), leading to inflated performance metrics. When faced with new, unseen data, their performance drops significantly.
2. Memorization Over Understanding: Larger models like Llama-3-70B show a greater drop in accuracy, suggesting they rely more on memorization than genuine comprehension. For instance, Llama-3-70B's accuracy fell from 76.6% to 69.7% with generic-to-brand swaps.
Key Take-Away
The RABBITS dataset sheds light on a critical weakness in current language models' handling of drug names. While LLMs hold great promise, their ability to accurately interpret and respond to drug-related queries still needs significant improvement. Our research underscores the importance of robustness in AI for healthcare to ensure safe and reliable patient support.
Visit the RABBITS leaderboard on Huggingface to see how different models compare!